5 Minutes
Breakthrough: a map of plant stem-cell regulators
Plant stem cells drive organ formation and growth in crops, underpinning global food, feed and biofuel supplies. On September 16, 2025, researchers at Cold Spring Harbor Laboratory (CSHL) published a detailed gene expression atlas that locates rare stem-cell regulators in maize and Arabidopsis. Using single-cell RNA sequencing and microfluidics, the team mapped two classic regulators—CLAVATA3 and WUSCHEL—across thousands of shoot cells and discovered additional genes that appear conserved across species. This work creates a functional roadmap to link stem-cell activity with traits such as maize ear size and overall productivity.
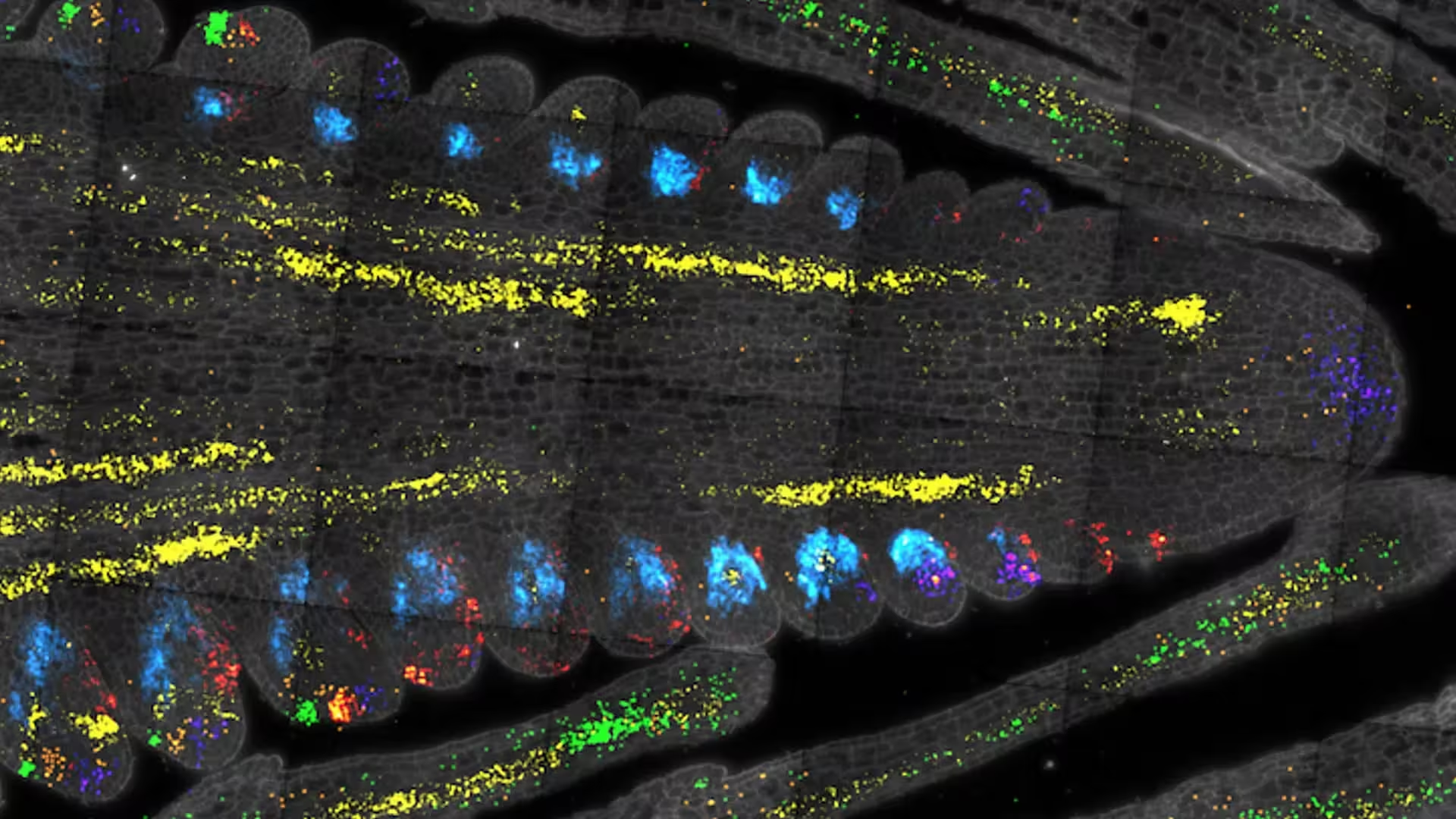
A thin section of a maize ear at a very early stage of development (about 3 millimeters long). Each color represents the expression of a different gene in stem cells and associated cells. Credit: Jackson lab/CSHL
Scientific background and why it matters
Plant stem cells reside in meristems, localized tissues that continuously supply new cells for leaves, stems and reproductive organs. The balance of stem-cell maintenance and differentiation determines organ size and yield. Despite their importance, stem cells are rare, embedded in heterogeneous tissues, and historically hard to profile with conventional bulk genomics. Identifying the genes that maintain stem-cell identity—the so-called "master switches"—could allow breeders and biotechnologists to design plants that regenerate more efficiently, produce larger harvest organs, or tolerate stress while maintaining yield. That has direct implications for food security and renewable biofuel production.
Methods: single-cell RNA sequencing and microfluidics
CSHL researchers combined careful microdissection with a droplet-based microfluidics pipeline. A former postdoc in David Jackson's lab, Xiaosa Xu, isolated tiny shoot segments from maize and Arabidopsis that contain meristematic stem cells. Each cell was separated by microfluidics, its mRNA reverse-transcribed into cDNA, and then barcoded so researchers could trace transcripts back to single cells. Single-cell RNA sequencing (scRNA-seq) reveals gene-expression patterns cell-by-cell rather than as averages across tissues, enabling recovery of rare cell populations.
Using this approach, the team recovered roughly 5,000 cells expressing CLAVATA3 and about 1,000 expressing WUSCHEL. From those cells they generated a gene-expression atlas that highlights hundreds of genes preferentially active in stem cells in both species—many of which are likely evolutionarily conserved regulators.
Key discoveries and implications
The atlas confirms the spatial and transcriptional signatures of known regulators and uncovers additional candidates linked to stem-cell identity. Importantly, some newly identified regulators show correlations with maize ear size and other productivity traits. Those genotype–phenotype links suggest a path to breed or engineer crops with optimized meristem activity to increase yield, improve organ uniformity, or adjust biomass for biofuel feedstocks.
CSHL Professor David Jackson commented on the broader potential: "Ideally, we would like to know how to make a stem cell. It would enable us to regenerate plants better. It would allow us to understand plant diversity. One thing people are very excited about is breeding new crops that are more resilient or more productive. We don't yet have a full list of regulators -- the genes we need to do that." The research team also emphasized that publishing the atlas creates an open resource for the community, reducing duplication of effort and accelerating follow-up work.
Related technologies and future prospects
This study illustrates how scRNA-seq and microfluidic technologies are transforming plant developmental biology. As single-cell techniques become more affordable and scalable, similar atlases can be produced across crop families and stress conditions, enabling comparative genomics of meristems. Combined with genome editing (CRISPR) and marker-assisted breeding, identified regulators could be validated and deployed to develop varieties tailored for higher yield, drought tolerance, or increased biomass for sustainable biofuels.
Expert Insight
Dr. Elena Moreno, plant geneticist and science communicator, offers context: "This atlas is a pivotal resource. By pinpointing rare stem-cell states and their regulators, the study moves us from descriptive genetics to actionable targets. For breeders, the immediate value is in markers linked to ear size and productivity; for molecular biologists, it provides candidate genes to test by editing or expression modulation. It's a clear example of how single-cell genomics bridges basic developmental biology and applied crop improvement."
Conclusion
The CSHL gene-expression atlas represents a major step toward decoding the regulatory networks that govern plant stem cells. By mapping conserved stem-cell regulators and connecting them to measurable traits in maize, the work provides a foundation for breeding and biotechnological strategies aimed at resilient, high-yield crops and optimized biomass for biofuels. As the atlas and associated datasets become publicly available, researchers across development, physiology and breeding will be able to build on these findings to reshape how we grow food and renewable plant-based resources.
Source: sciencedaily


Leave a Comment